μCaler FS EMS+ Panel v1.0
#1101441
μCaler FS EMS+ Panel v1.0 includes selected sites approved by the NMPA and FDA, as well as those reported in literature and patents, totaling 1,783 CpG sites. It covers 10 major high-incidence cancers, including lung cancer, breast cancer, colorectal cancer, prostate cancer, gastric cancer, liver cancer, cervical cancer, bladder cancer, esophageal cancer, and ovarian cancer, involving 163 methylated candidate genes associated with carcinogenesis and tumor suppression. The panel’s target regions cover approximately 0.1 Mb of the human genome, with 9,680 CpG sites, providing strong technical support for comprehensive and accurate methylation early screening and MRD recurrence monitoring.
- Product details
- Kit Content
- Ordering Information
- Resources
Feature
- Sample Compatibility: Suitable for gDNA, cfDNA, and various level of FFPE samples.
- Multi-cancer Detection: Covers large-scale CpG sites of multiple genes for 10 major high-incidence cancers worldwide in a single detection.
- Flexible Customization: Supports tailored probe sets with various CpG site scales and/or gene mutations for single or multiple cancer types, enabling synchronous detection of methylation and mutations.
- Accurate Quantification: Achieves precise quantification of methylation levels (0 ~ 100%) in tested samples.
- Efficient Capture: Requires no sequence conversion and retains native sequence complexity, increasing effective coverage depth by several times.
-
Ultra-high Sensitivity: Compatible with unique dual indexed adapters including unique molecular identifier (UMI) for ultra-sensitive detection of low to ultra-low abundance signals.
Performance
Accurate Quantification of Methylation Levels
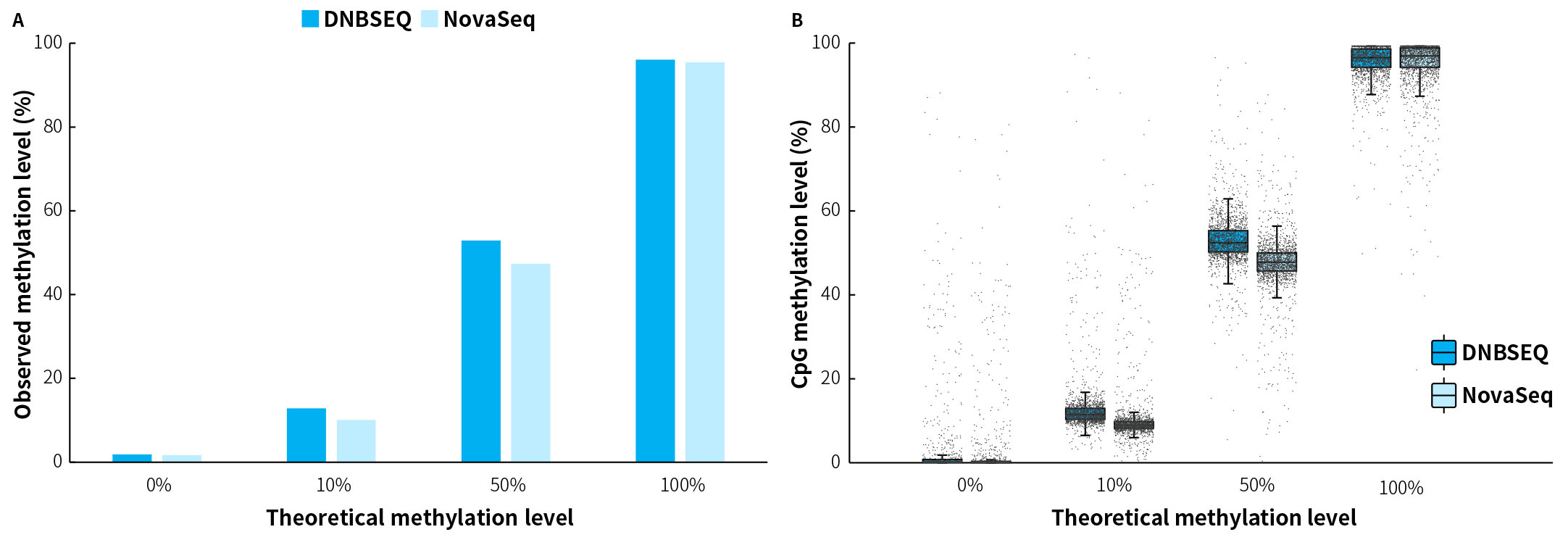
Figure 1. Accurate quantification of simulated samples with different methylation levels by μCaler FS EMS+ Panel v1.0. A. Comparison between theoretical and observed methylation levels; B. Methylation levels of CpG sites in target regions. Simulated samples were processed to hybridization capture using the μCaler DNA Full Screen System and μCaler FS EMS+ Panel v1.0. Sequencing was performed on DNBSEQ-T7 (PE150) and NovaSeq 6000 (PE150). For each sample, 1 M reads pair were randomly selected for data analysis.
Note: The samples consisted of 100% Methylated DNA (Zymo, D5014-2) as a positive control and 0% Methylated DNA (Zymo, D5014-1) as a negative control, mixed at different ratios to simulate methylation levels (0%, 10%, 50% and 100%). The initial input amounts were 50 ng.
Maintaining High Stability in Methylation Detection
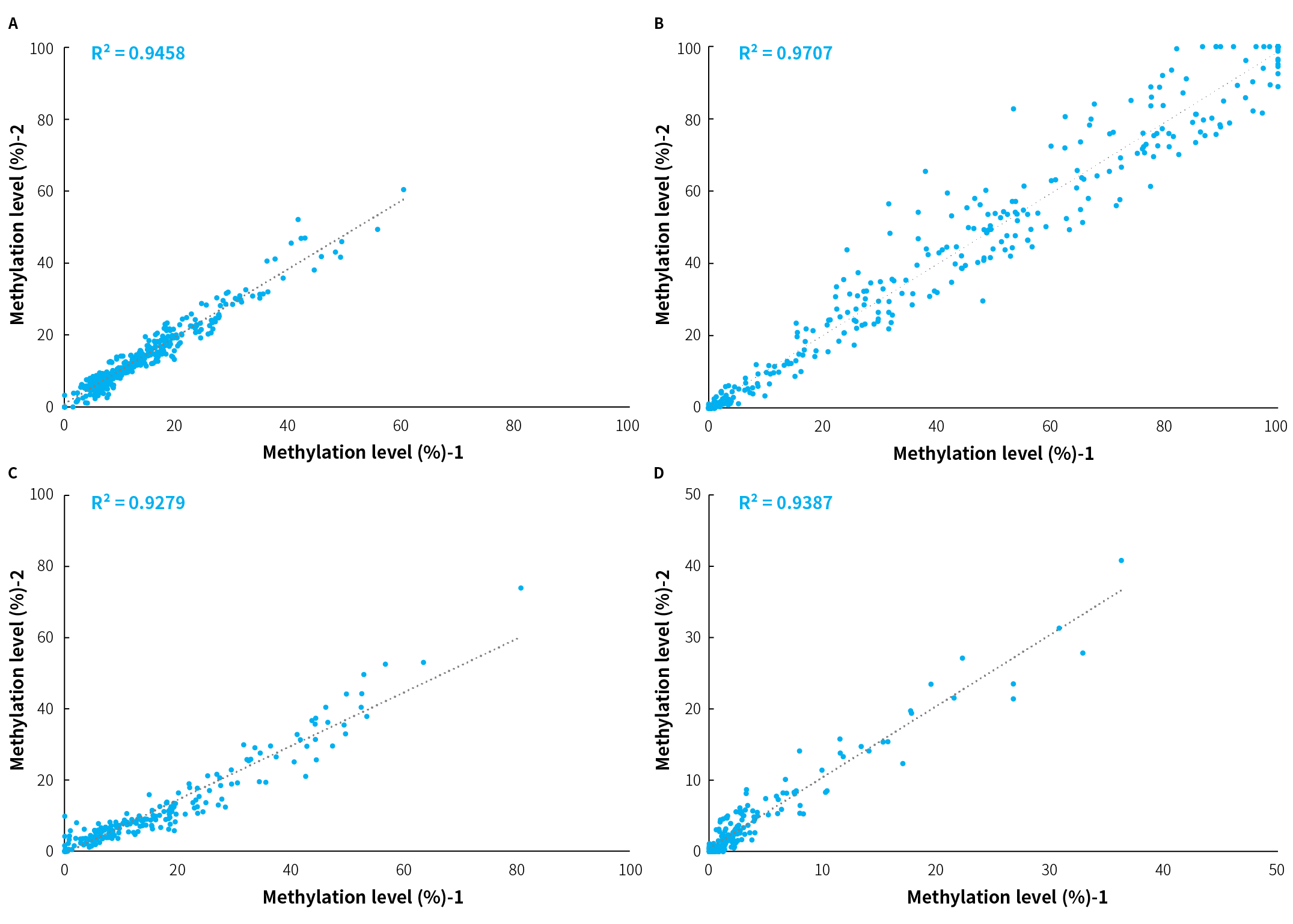
Figure 2. Correlation analysis of methylation levels detected between two repeats of the sample by μCaler FS EMS+ Panel v1.0. A-D. The correlation of methylation levels between two replicates of 4 independent samples. 4 clinical tumor tissue samples were randomly selected (A & B: colorectal cancer, C & D: bladder cancer) and processed to hybridization capture using the μCaler DNA Full Screen System and μCaler FS EMS+ Panel v1.0. The R² values for correlation ranged from 0.9279 to 0.9707.
Note: The initial input amounts were 20 ng.
Effectively Distinguishing Tumor and Peritumoral Tissues
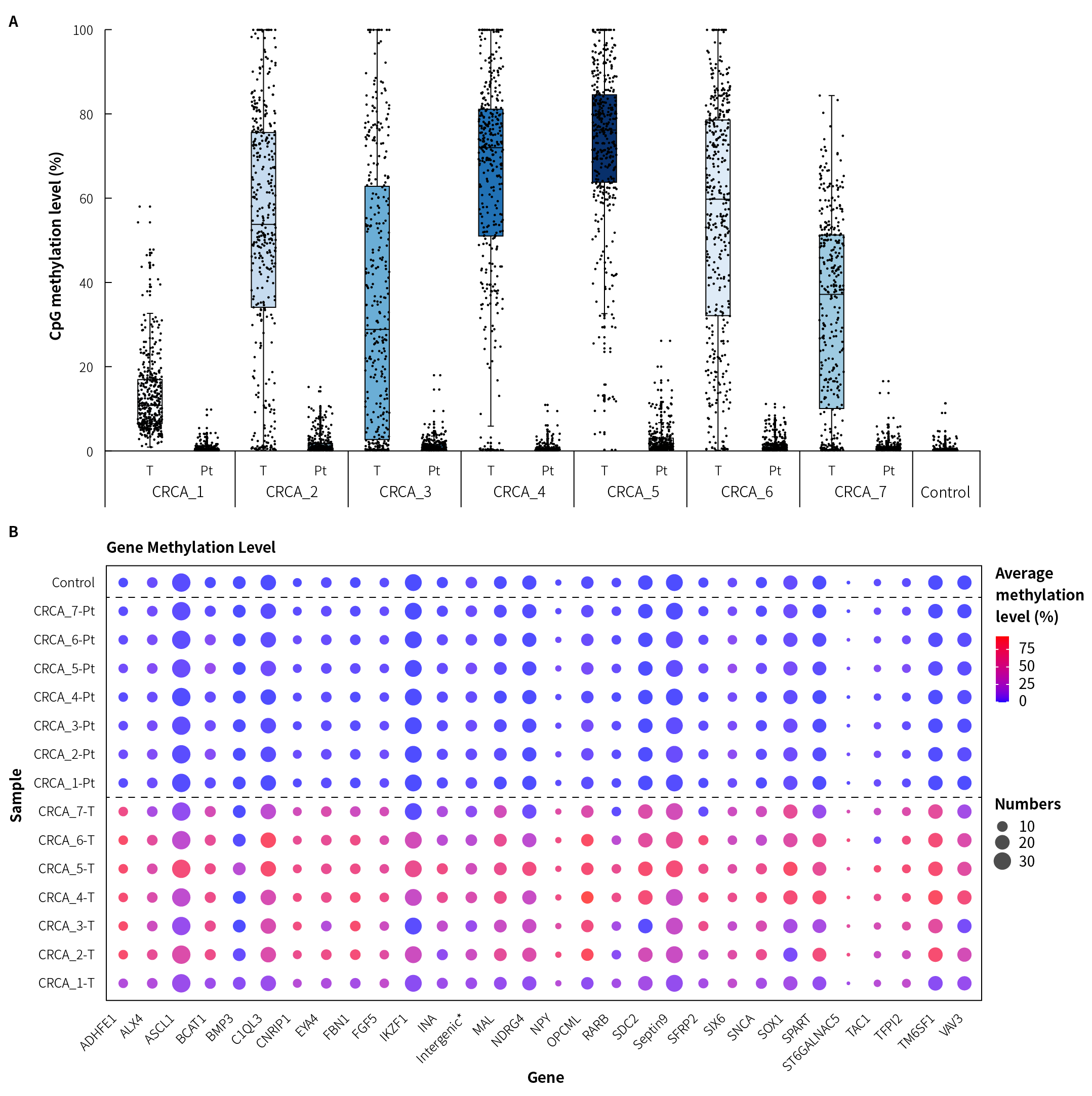
Figure 3. Methylation level detection of μCaler FS EMS+ Panel v1.0 applied to clinical colorectal cancer and peritumoral tissue samples. A. CpG methylation levels in paired colorectal cancer samples;B. CpG methylation levels of colorectal cancer candidate biomarkers in clinical samples.
Note: CRCA: Colorectal Cancer; Pt: Peritumoral tissue. Control refers to Human Genomic DNA standard (Promega, G1471), with an input of 20 ng.
Genelist
Type of cancer
Biomarkers
Lung cancer
BCAT1
CDH1
CDKN2A
CDO1
DMRTA2
FOXD3
FOXI2
GATA5
GDNF
HOXA7
HOXA9
OPCML
PAX5
PTGER4
RASSF1
SCT
SHOX2
SIX6
SLC12A8
SOX17
ST6GALNAC5
SULF2
TAC1
TNFRSF25
ZFP42
Breast cancer
APC
ASCL1
BRCA1
C1QL3
CPXM1
DKK3
GALR1
HOXA10
HOXA9
ITIH5
LBX1
OTP
PITX2
RARB
RASGRF1
SOX1
USP44
Colorectal cancer*
ADHFE1
ALX4
ASCL1
BCAT1
BMP3
C1QL3
CNRIP1
EYA4
FBN1
FGF5
IKZF1
INA
MAL
NDRG4
NPY
OPCML
RARB
SDC2
SEPTIN9
SFRP2
SIX6
SNCA
SOX1
SPART
ST6GALNAC5
TAC1
TFPI2
TM6SF1
VAV3
Prostate cancer
AKR1B1
ANXA2
AOX1
APC
CFAP90
GDAP1L1
GSTP1
HAAO
PRKCB
PTGS2
RARB
RUNX3
SEPTIN9
SIX6
TAC1
Gastric cancer
ADRA1B
ARHGAP20
ASCL1
C1QL3
CABIN1
CD40
CPEB1
DLGAP3
DOCK10
ELMO1
HOPX
HOXD11
KCNQ5
LMX1A
RNF180
RORB
RPRM
RUBCNL
RUNX3
SDC2
SEPTIN9
TCF4
TFPI2
ZNF569
Liver cancer
ALX3
ARHGEF33
C1QL3
CDKL2
CFTR
DCDC2
EVX1
FAR1
HOXA11
HOXA9
MTHFD2
PLAC8
RIPK3
RUNX3
SEPTIN9
SGIP1
SH3YL1
SIX6
SOX1
TAMALIN
TBX15
WNT3A
ZNF605
Cervical cancer
ASCL1
ASTN1
C1QL3
CHAD
CHRM2
DLX1
EDN3
EPB41L3
FGF10
HOXA9
ITGA4
ITGA8
JAM3
OPCML
PAX1
PCDHGB7
RXFP3
SEPTIN9
SIX6
SOX1
SOX17
ST6GALNAC5
TAC1
TAFA4
TFPI2
ZNF582
ZNF781
ZSCAN1
Bladder cancer
EOMES
FEZF2
HAND2
IRAK3
IRX4
LRRC4
NID2
NOVA1
NRN1
ONECUT2
OTX1
PENK
PIGG
SOX1
TAC1
TCERG1L
TMEFF2
TWIST1
VIM
Esophaheal cancer
ASCL1
CCNA1
EPO
HOXC10
HOXD1
KCNA3
MAFB
MT1A
MT1G
OPCML
RAB37
RAPGEFL1
SEPTIN9
SIX6
ST3GAL6
ST6GALNAC5
TAMALIN
TFPI2
ZNF132
ZNF154
ZNF582
Ovarian cancer
CDH13
CDO1
GYPC
HNF1B
HOXA11
HOXA9
IFFO1
LOC284798
LYPD5
OPCML
PAX1
PCDH17
PCDHB18P
RASSF1
RYBP
SOX1
TAC1
ZNF671
*It also covers intergenic regions not listed in the table (Intergenic, containing 12 CpG sites).
For research use only. Not for use in diagnostic procedures.
| Catalog# | Cap Color | Item | Volume | Package/Storage |
| 1101442 |
|
μCaler FS EMS+ Panel v1.0, 16 rxn | 40 μL | -25~-15℃ |
| 1101441 |
|
μCaler FS EMS+ Panel v1.0, 96 rxn | 210 μL | -25~-15℃ |
| Product | Catalog# |
| μCaler FS EMS+ Panel v1.0, 16 rxn | 1101442 |
| μCaler FS EMS+ Panel v1.0, 96 rxn | 1101441 |
Product Sheet
-
[Product Catalog] -
Product Catalog
Download
-
[Poster] -
EN-Brochure-μCaler FS EMS+ Panel v1.0
Download
-
[Technical Note] -
EN-Instruction-μCaler FS EMS+ Panel v1.0
Download
-
[Technical Note] -
EN_Protocol_μCaler_DNA_Full_Screen_System_Comprehensive_Solution_for_Illumina®_V1.0
Download
-
[Technical Note] -
EN_Protocol_μCaler_DNA_Full_Screen_System_Comprehensive_Solution_for_MGI_V1.0
Download
Application Note
Demo Data
Solutions
- Methyl Library Preparation Total Solution
- Sequencing single library on different platform--Universal Stubby Adapter (UDI)
- HRD score Analysis
- Unique Dual Index for MGI platforms
- RNA-Cap Sequencing of Human Respiratory Viruses Including SARS-CoV-2
- Total Solution for RNA-Cap Sequencing
- Total Solution for MGI Platforms
- Whole Exome Sequencing
- Low-frequency Mutation Analysis
Events
-
Exhibition Preview | Nanodigmbio invites you to join us at Boston 2025 Annual Meeting of the American Society of Human Genetics (ASHG)

-
Exhibition Preview | Nanodigmbio Invites You to Join Us at WHX & WHX Labs Kuala Lumpur 2025, Malaysia International Trade and Exhibition Centre in Kuala Lumpur

-
Exhibition Preview | Nanodigmbio Invites You to Join Us at Hospitalar 2025, Brazil International Medical Device Exhibition in São Paulo

-
Exhibition Preview | Nanodigmbio invites you to join us at Denver 2024 Annual Meeting of the American Society of Human Genetics (ASHG)

-
Exhibition Preview | Nanodigmbio invites you to join us at Sapporo 2024 Annual Meeting of the Japan Society of Human Genetics (JSHG)

-
Exhibition Preview | Nanodigmbio invites you to join us at Association for Diagnostics & Laboratory Medicine (ADLM)

Nanodigmbio will contact you within 1 day.
If there are any problems, please contact us by 400 8717 699 / support@njnad.com